The first step of CRISPR/Cas9 gene editing is designing a single guide RNA (sgRNA) to target your gene of interest. Because sgRNAs are solely responsible for recruiting Cas9 to specific genomic loci, optimal sgRNA design is critical for successful gene editing experiments. There are many web-based tools available for sgRNA design, each of which has different features and advantages. The information provided here will help you choose the best tool for your specific research objective.
Several web-based tools available
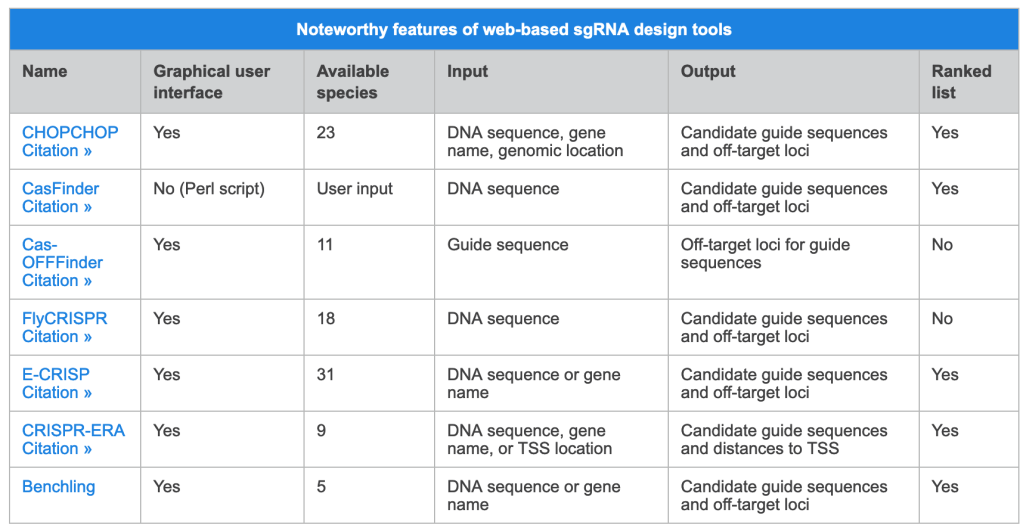
Web-based sgRNA design tools typically require that users input a DNA sequence, genomic location, or gene name for each gene of interest, and indicate a species. An algorithm specific to each tool outputs a list of candidate guide sequences with corresponding predicted off-target sites for each input (Wu et al. 2014). Most tools aim to provide guide sequences that minimize the likelihood of off-target effects, but the methods they employ vary. For example, Chop Chop uses empirical data from multiple recent publications (e.g. Doench et al. 2016) to calculate efficiency scores. Alternatively, CasFinder (Aach et al. 2014) and E-CRISP (Heigwer et al. 2014) incorporate specific user-defined penalties based on the number and position of mismatches relative to the guide sequence in order to rank the potential for off-target effects.
Tools for specific applications
Some sgRNA design tools have been developed for specific applications. CRISPR-ERA (Liu et al. 2015) is the only currently available tool that designs sgRNAs specifically for gene repression or activation, while FlyCRISPR (Gratzet al. 2013) focuses on applications in fly, beetle, and worm species, including the popular model organisms Drosophila melanogaster and Caenorhabditis elegans. Presently, the design tool featured on the Benchling website is the only one that can generate candidate sgRNAs that are compatible with alternative nucleases such as Staphylococcus aureus Cas9 (Ran et al. 2015) and Cpf1 (Zetsche et al. 2015). Given the uniqueness of each tool, we recommend that you use multiple approaches during the sgRNA design process and choose guide sequences that are consistently predicted to perform well.
A selection of freely available tools
The table below provides a list of web-based tools for sgRNA design. For simplicity, we have only included those that are free and do not require a subscription. For each tool, we have indicated whether there is a convenient graphical user interface or if the user has to download a script. If you would prefer to design sgRNAs manually, please visit Choosing a target sequence for CRISPR/Cas9 gene editing to learn more.