How did “genome editing” become a household phrase so quickly? This question, posed by Jerel Davis of the investment firm Versant Ventures, opened a gene-editing panel at the 2019 Life Science Innovation Northwest (LSINW) conference in Seattle, Washington. “Genome editing is a juxtaposition of two discoveries,” explained panelist Philip Gregory from the gene and cell therapy company Bluebird Bio: Nucleases can make double-stranded DNA breaks (DSBs) at specific sequences, and DSBs activate repairs that can change DNA.
DSB repair has two mechanisms. Nonhomologous end joining (NHEJ) links ends together, often creating insertions and deletions (indels) in the process. In genome editing, this can be used to knock out gene function. Homology-directed repair (HDR) fixes DSBs using DNA with a similar sequence. Providing cells with external homologous donor DNA introduces edits via HDR.
Many genome-editing systems work by activating DSB repair at specific sites using engineered zinc-finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALENs), or meganucleases (1). Currently, the dominant genome-editing method is CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats-CRISPR-associated protein 9) (2). How do researchers choose among these systems?
“The primary consideration is the end product,” says Jon Hennebold, Oregon Health & Science University in Portland. Hennebold leads a multisite U.S. National Institutes of Health–funded program on genome-editing efficiency and safety. Companies use proprietary genome-editing systems optimized for specificity to reduce off-target effects (mutations at unintended sites). Most academic labs can get the product they want with CRISPR, which is fast and easy. “You can order the components and get started in 24 to 48 hours,” Hennebold says, “Other methods don’t have that commercial support.”
CRISPR: It gets the job done
Academic labs have no reason to work with other methods,” says Charles Gersbach, a biomedical engineer at Duke University in Durham, North Carolina. “For plain-vanilla genome editing, Cas9 and a gRNA will get the job done.” Cas9, an enzyme from bacterial antiviral systems, makes DSBs at DNA sites that are complementary to a guide RNA (gRNA) and also have a nearby protospacer-adjacent motif (PAM) sequence. CRISPR repeats aren’t needed for editing, so Cas9 plus a gRNA can knock out genes by NHEJ. Providing a DNA fragment promotes HDR-mediated edits.
Ru Gunawardane, director of Stem Cells and Gene Editing at Seattle’s Allen Institute for Cell Science and an LSINW panelist, says CRISPR has been “a game changer” in fulfilling the institute’s mission of understanding how cells act in normal, disease, and treatment conditions. Researchers at the institute use CRISPR to tag organelle markers in stem cells with fluorescent proteins, then track these fusion proteins and their interactions under different situations. Currently, their work includes differentiating tagged cells into cardiomyocytes.
“We’ve tagged 40 to 50 sites so far,” Gunawardane says. “Once you have the CRISPR platform, all you have to change is the gRNA and the template for introducing the tag at the right location in the genome.” However, institute researchers do months of downstream quality control, such as live imaging and sequencing, before using the cells experimentally or making them available for research.
Caixia Gao, a plant biologist at the Chinese Academy of Sciences in Beijing, says CRISPR is also common in her field. “All methods are very efficient at making site-specific mutations,” she says, “but CRISPR takes the least time and has the lowest costs.”
CRISPR alternatives
CRISPR-mediated genome editing has drawbacks, though. The PAM requirement limits target sequences. Cas9 is large, so its gene is difficult to deliver to cells via vectors such as adeno-associated viruses commonly used in gene therapy. Scientists worry about off-target effects, although experts note that concerns about unintended mutations are often based on calculations from studies on improving editing. These studies may deliberately use low-specificity conditions to facilitate monitoring progress.
To ensure the highest confidence in their products, companies invest time and money in custom genome-editing methods focused on efficiency and specificity. Initial investments pay off, industry scientists say, by preventing problems later in development.
ZFNs are the genome-editing reagents used by the genomic medicine company Sangamo, based in Brisbane, California. Chief Technology Officer Ed Rebar explains that Sangamo’s core editing reagent is a ZFN dimer. The typical target site is 36 basepairs. Each ZFN is a chimeric protein of the nuclease domain from the FokI restriction enzyme and an array of zinc-finger DNA-binding domains built by “mixing and matching” from Sangamo’s archive of thousands of two-finger subunits. Strategies for diversifying the ZFN architecture for high targeting capability include attaching the FokI domain to the N- or C-terminus of the zinc-finger array and inserting base-skipping linkers between fingers. With Sangamo’s high-throughput, automated process for generating ZFNs, Rebar says, “Starting from a target gene name, we can generate an initial set of editing reagents within two weeks.”
In a demonstration study, Rebar and colleagues designed ZFNs that introduced indels at 25 of 28 bases in a promoter relevant to studying hemoglobinopathies (3). Despite this precision and the advantage of being smaller than Cas9, ZFNs are not as commonly used as CRISPR-based methods. Sangamo provides ZFNs via industry and academic partnerships but holds the modules, expertise—and patents—for making them.
TALENs attach FokI to arrays of DNA-binding modules, originally from plant pathogens, that each target a single basepair. TALENs are smaller than Cas9, but larger than ZFNs. The modules have high DNA-binding affinity but include repeated sequences that create cloning challenges.
Dan Carlson is chief scientific officer at Recombinetics, a St. Paul, Minnesota–based biotechnology company that uses TALENs and CRISPR to generate animals and cell lines for clinical research models and agriculture. Using these methods, he says, “we can target almost any site in a genome.” With in-house resources, even TALENs take only “a few hundred bucks and about a week” to generate, Carlson adds, so scientists choose the method that is most reproducible, consistent, and specific, based on pilot studies. These initial investments ensure the company is responsible with resources, he says. “It costs too much to sort out problems on the back end.”
Meganucleases, also called homing endonucleases, are smaller than Cas9, despite their name, which refers to recognition sequences that can be up to 40 basepairs in length. Hybrid megaTALs combine the simple assembly of TALENS with the DNA-cleavage specificity of meganucleases. Two biotech companies that use meganuclease-based methods are Bluebird Bio in Cambridge, Massachusetts, and Precision BioSciences in Durham, North Carolina.
Barry Stoddard, a structural biologist at Fred Hutchinson Cancer Research Center, Seattle, has a panel of 50–60 meganucleases that his lab engineers to recognize specific sequences. “It takes one day to make CRISPR to target a gene,” he says, “and 100 days to make a meganuclease.” Still, Stoddard gets many requests for engineered meganucleases, because their precision is highly valued for applications such as developing therapeutics for which “100 days is nothing.”
To DSB or not to DSB
Relying on HDR for editing risks introducing indels or chromosomal translocations. Even with a precisely targeted nuclease, with HDR, “you’re at the mercy of the cell,” Stoddard observes. For editing without the unpredictability of HDR, he adds, watch for developments in site-specific recombinases (SSRs).
“SSRs do the whole thing,” says Marshall Stark, molecular geneticist at the University of Glasgow in Scotland. “They break and rejoin the DNA with no need for host factors.” Even in cells with low or no HDR, SSRs can integrate exogenous DNA at a targeted site. Under optimal conditions, Stark says, “SSRs can be extremely efficient, with recombination approaching 100% in a few minutes.” SSRs can make switch-like changes such as inverting a DNA segment’s orientation. This makes them valuable for creating electronic circuit–like pathways that control cell behavior for industrial purposes or synthetic biology applications, such as making biocomputers. However, SSRs have complex, rare, 30- to 50-basepair target sites.
Stark names two approaches to adapting SSRs for more widespread genome editing: (1) using directed evolution that selects for new target specificities and (2) making fusion proteins. For example, he and others are attaching SSR-derived recombinase domains to zinc-finger modules that bind specific DNA sequences. The technology is “still at the investigative level,” Stark notes. “If you have a particular target in mind, it’s still a lot of work to make a recombinase for it.”
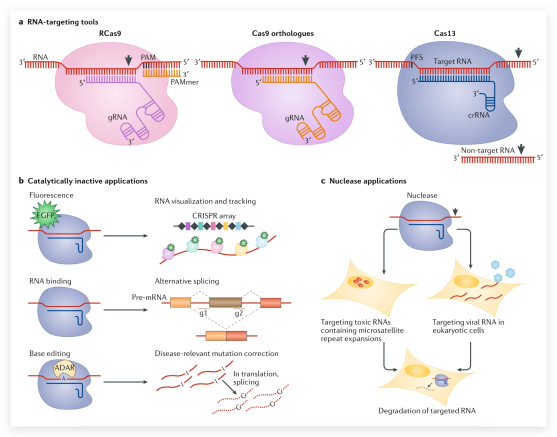
For genome-editing without DSBs, researchers use Cas9 that is still directed by gRNAs but does not cut DNA or makes only single-stranded nicks. Cas9 variants are fused to transcription activators or repressors, or to enzymes that alter chromatin structure by modifying DNA or DNA-packaging histone proteins to change gene expression. This epigenome editing resembles natural gene regulation, Gersbach says, “without risk of off-target changes to DNA sequences.” The method is a basic research tool for studying epigenetics and has potential therapeutic uses, such as reactivating the silenced gene that causes the intellectual disability Fragile X syndrome (4).
Base editing makes single-basepair changes while avoiding unintended mutations from DSB repair. It works in cells without HDR. Innovations in this method are published regularly, but the first base editors developed by David Liu’s group at Harvard University had a disabled Cas9 targeting a DNA sequence fused to an enzyme that converts cytosine to uracil. The fusion protein changes a cytosine–guanine pair to thymine–adenine. Another base editor, which Liu’s lab generated through protein engineering and directed evolution, changes adenine–thymine to guanine–cytosine. Just these two base-editing systems can make one-third of all possible basepair changes, Liu asserts, and potentially correct 62% of known human pathogenic point mutations.
As of summer 2019, more than 100 research papers described experiments using base editing, Liu says, “including several that corrected animal models of human genetic diseases by directly reversing point mutations.” For example, one editor corrected a mutation that causes phenylketonuria (5). Liu and others are diversifying base editing—expanding base-changing options, increasing specificity, and improving activity in live animals and at target sites that require distinguishing between highly similar sequences.
Our genome-editing future
As a scientist using genome-editing technology, Carlson hopes researchers apply it for the good of humankind and the planet. He hopes the public understands that getting a final product is actually a long process. The biotechnology company Recombinetics got media attention for using TALENs to breed polled (hornless) cows—which saves farmers the trouble of dehorning them. The project started in 2012, and Carlson says the company continues to work on making the editing more efficient.
Given its popularity and availability, CRISPR dominates genome-editing predictions. CRISPR-based systems will continue to improve incrementally, Carlson says. Researchers regularly publish about improved gRNAs with higher efficiency or specificity. Multiple Cas-type enzymes have been discovered or engineered with different PAMs or activities (6). For example, Cas13 targets RNA and is the foundation of RNA base editing. This method and Liu’s DNA base editing are licensed to Beam Therapeutics, whose cofounders include Liu and Feng Zhang, who developed CRISPR for mammalian cells.
CRISPR methodological improvements include treating cells with small molecules during editing to nudge DSB repair away from NHEJ and toward HDR. Controllable systems switch on Cas9 using light or small molecules, limiting its activity in order to reduce off-target effects. Researchers are scouring the microbial world for new Cas-type enzymes and entirely new genome-editing systems. “We’re still identifying new molecules with editing capacity and we don’t fully understand the editing tools we have,” Hennebold says. “We still have a lot to learn.”
The practice of using CRISPR to correct disease-causing mutations is growing: Editas Medicine and Allergan announced human in vivo CRISPR-therapy trials for an inherited blindness. A potential hurdle to therapeutic CRISPR is the possibility of human immune responses to its bacterial components. For instance, a majority of tested blood samples showed existing immune responses to Cas9, which is commonly taken from Staphylococcus or Streptococcus bacteria (7).
The genome-editing wish list includes better methods for multiplexing—editing more than one gene at a time. For example, multiplexing would speed developments in T-cell–based immunotherapy, which works for many patients but requires altering multiple genes. And plant scientists often want to create “stacks” of linked genes that are inherited together as a package for resistance to disease, pests, and other agricultural threats. Multiplexing would accelerate creating these products.
In principle, multiplexing is simple with CRISPR, requiring only the introduction of a single Cas enzyme and of gRNAs and template DNAs for each targeted gene. Gunawardane has tried CRISPR multiplexing to tag multiple genes in the same cell and says it’s achievable, but in practice, gets increasingly complicated with each added gene. Systems using SSRs, ZFNs, or meganucleases may offer advantages such as smaller components that allow easier introduction.
Ask scientists about genome-editing challenges and they mention delivery of components into cells. They say to watch for transient systems that deliver editing enzymes as proteins instead of their genes so that the proteins are degraded after acting instead of being continuously expressed. Limiting activity in this manner could reduce off-target effects. Gao notes that DNA-independent delivery of genome-editing systems could alleviate concerns about genetically modified organisms (GMOs). “Proteins can’t integrate into the genome,” she says, “so if no foreign DNA is delivered at all, the resulting plants should be considered non-GMO.”
CRISPR is already very powerful, and so many people are working on it and other genome-editing systems that they’ll inevitably continue to improve, Gao says. “Scientists like to make new tools and new technology,” she says, “so we’re really seeing progress every day. Now, we say we can edit any target in principle, but in five years it will be true.”
References
A. J. Bogdanove, et al., Nucleic Acids Res. 46, 4845–4871 (2018), doi: 10.1093/nar/gky289.
A. C. Komor, A. H. Badran, D. R. Liu, Cell 168, 20–36 (2017), doi: 10.1016/j.cell.2016.10.044.
D. E. Paschon et al., Nat. Comm. 10, 1133 (2019), doi: 10.1038/s41467-019-08867-x.
X. S. Liu et al., Cell 172, 979–992 (2018), doi: 10.1016/j.cell.2018.01.012.
L. Villiger et al., Nat. Med. 24, 1519–1525 (2018), doi: 10.1038/s41591-018-0209-1.
A. Pickar-Oliver, C. A. Gersbach, Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019), doi: 10.1038/s41580-019-0131-5.
C. T. Charlesworth et al., Nat. Med. 25, 249–254 (2019), doi: 10.1038/s41591-018-0326-x.
Source: https://www.sciencemag.org/features/2019/09/beyond-crispr-what-s-current-and-upcoming-genome-editing