“Proximity-CLIP can be used to simultaneously map the compartment-specific landscape of RBPs, the transcriptome and RBP-occupied RNA loci”
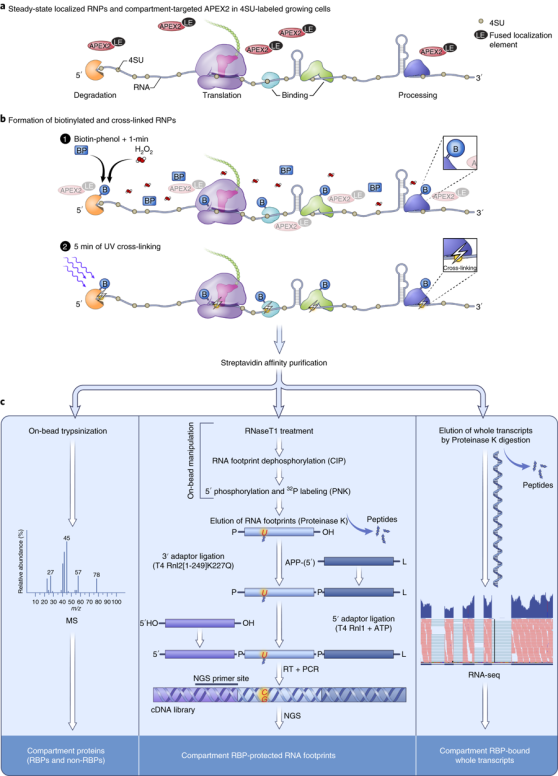
The spatial compartmentalization of RNA is pivotal to gene expression and its regulation. However, the study of RNA localization has been hampered by a lack of high-throughput methods for mapping the transcriptomes of different cellular compartments. A new method called Proximity-CLIP combines proximity-labelling of proteins with ultraviolet (UV) crosslinking and next-generation sequencing to map the subcellular locations of RNAs.
The team built on the previously established APEX-RIP, a technique for the high-throughput mapping of RNA localization in intact cells that combines compartment-specific protein biotinylation with protein–RNA formaldehyde crosslinking. The engineered peroxidase APEX2 functions as a labelling tag that is genetically targeted to subcellular compartments through fusion with compartment-specific localization signals. Addition of biotin-phenol, a small-molecule substrate for APEX2, results in the biotinylation of endogenous proteins in close proximity to the enzyme. Proteins can subsequently be identified by mass spectrometry, whereas crosslinked RNAs can be identified by RNA sequencing (RNA-seq).
In Proximity-CLIP, RNAs are first labelled with 4-thiouridine (4SU) in living cells that express specifically localized APEX2. RNA and proteins are crosslinked in vivo by UV light, rather than by chemical crosslinking, during the quenching step that follows the biotinylation of APEX2-proximate proteins. Localized, biotinylated and crosslinked ribonucleoprotein complexes are then isolated using streptavidin affinity chromatography. While the proteome is determined using mass spectrometry, the RNA can either be analysed by standard RNA-seq to identify local transcripts or undergo RNase treatment to reveal RNase-resistant ‘footprints’, which reveal the occupancy of these sequences by RNA-binding proteins (RBPs). Sequencing of cDNA libraries generated from footprints enables the identification and quantification of RBP-occupied cis-acting regulatory elements on transcripts.
The UV crosslinking of RNA labelled with 4SU leads to a T-to-C mutation in the corresponding cDNA libraries. This mutation can be exploited to bioinformatically remove contaminating RNAs that bind nonspecifically to the affinity chromatography matrix, thereby increasing the specificity of Proximity-CLIP. UV crosslinking also avoids some pitfalls of formaldehyde crosslinking, such as the crosslinking of long-range or indirect interactions.
Applying their technique to HEK293 cells inducibly expressing APEX2 fused either to a nuclear export signal or to histone H2B, Benhalevy et al. determined both the proteome and transcriptome of the cytoplasm and nucleus, respectively. Functional enrichment analysis showed that proteins biotinylated by H2B-APEX2 were involved in transcription or belonged to nuclear categories, such as ‘nucleus’ and ‘nucleoplasm’. Moreover, the protein profiles resembled those previously reported in a study that used mass spectrometry to determine nuclear and cytoplasmic proteomes.
Transcriptome profiles of the cytoplasmic and nuclear compartments obtained using Proximity-CLIP matched RNA profiles obtained after biochemical fractionation. Furthermore, sequencing of RBP-protected fragments from both nucleus and cytoplasm revealed a footprint profile that reflected each compartment; that is, the vast majority of RBP footprints on mRNA in the cytoplasm were found on mature mRNA, whereas 43% of mRNA footprints in the nucleus resided in introns.
Taken together, Proximity-CLIP can be used to simultaneously map the compartment-specific landscape of RBPs, the transcriptome and RBP-occupied RNA loci. Its high throughput compared with imaging-based techniques makes it particularly appealing for identifying the subcellular localization of transcripts and potentially interacting RBPs.
【REF】
Benhalevy, D. et al. Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods 15, 1074–1082 (2018)
Koch L. Proximity-CLIP – close encounters of the RNA kind. Nat Rev Genet. 2019 Feb;20(2):68-69